CARP returns a fast approximation to the Convex Clustering
solution path along with visualizations such as dendrograms and
cluster paths. CARP solves the Convex Clustering problem via an efficient
Algorithmic Regularization scheme.
CARP( X, ..., weights = sparse_rbf_kernel_weights(k = "auto", phi = "auto", dist.method = "euclidean", p = 2), labels = rownames(X), X.center = TRUE, X.scale = FALSE, back_track = FALSE, exact = FALSE, norm = 2, t = 1.05, npcs = min(4L, NCOL(X), NROW(X)), dendrogram.scale = NULL, impute_func = function(X) { if (anyNA(X)) missForest(X)$ximp else X }, status = (interactive() && (clustRviz_logger_level() %in% c("MESSAGE", "WARNING", "ERROR"))) )
Arguments
| X | The data matrix (\(X \in R^{n \times p}\)): rows correspond to
the observations (to be clustered) and columns to the variables (which
will not be clustered). If |
|---|---|
| ... | Unused arguements. An error will be thrown if any unrecognized
arguments as given. All arguments other than |
| weights | One of the following:
|
| labels | A character vector of length \(n\): observations (row) labels |
| X.center | A logical: Should |
| X.scale | A logical: Should |
| back_track | A logical: Should back-tracking be used to exactly identify fusions? By default, back-tracking is not used. |
| exact | A logical: Should the exact solution be computed using an iterative algorithm?
By default, algorithmic regularization is applied and the exact solution
is not computed. Setting |
| norm | Which norm to use in the fusion penalty? Currently only |
| t | A number greater than 1: the size of the multiplicative update to
the cluster fusion regularization parameter (not used by
back-tracking variants). Typically on the scale of |
| npcs | An integer >= 2. The number of principal components to compute for path visualization. |
| dendrogram.scale | A character string denoting how the scale of dendrogram
regularization proportions should be visualized.
Choices are |
| impute_func | A function used to impute missing data in |
| status | Should a status message be printed to the console? |
Value
An object of class CARP containing the following elements (among others):
X: the original data matrixn: the number of observations (rows ofX)p: the number of variables (columns ofX)alg.type: theCARPvariant usedX.center: a logical indicating whetherXwas centered column-wise before clusteringX.scale: a logical indicating whetherXwas scaled column-wise before centeringweight_type: a record of the scheme used to create fusion weights
Examples
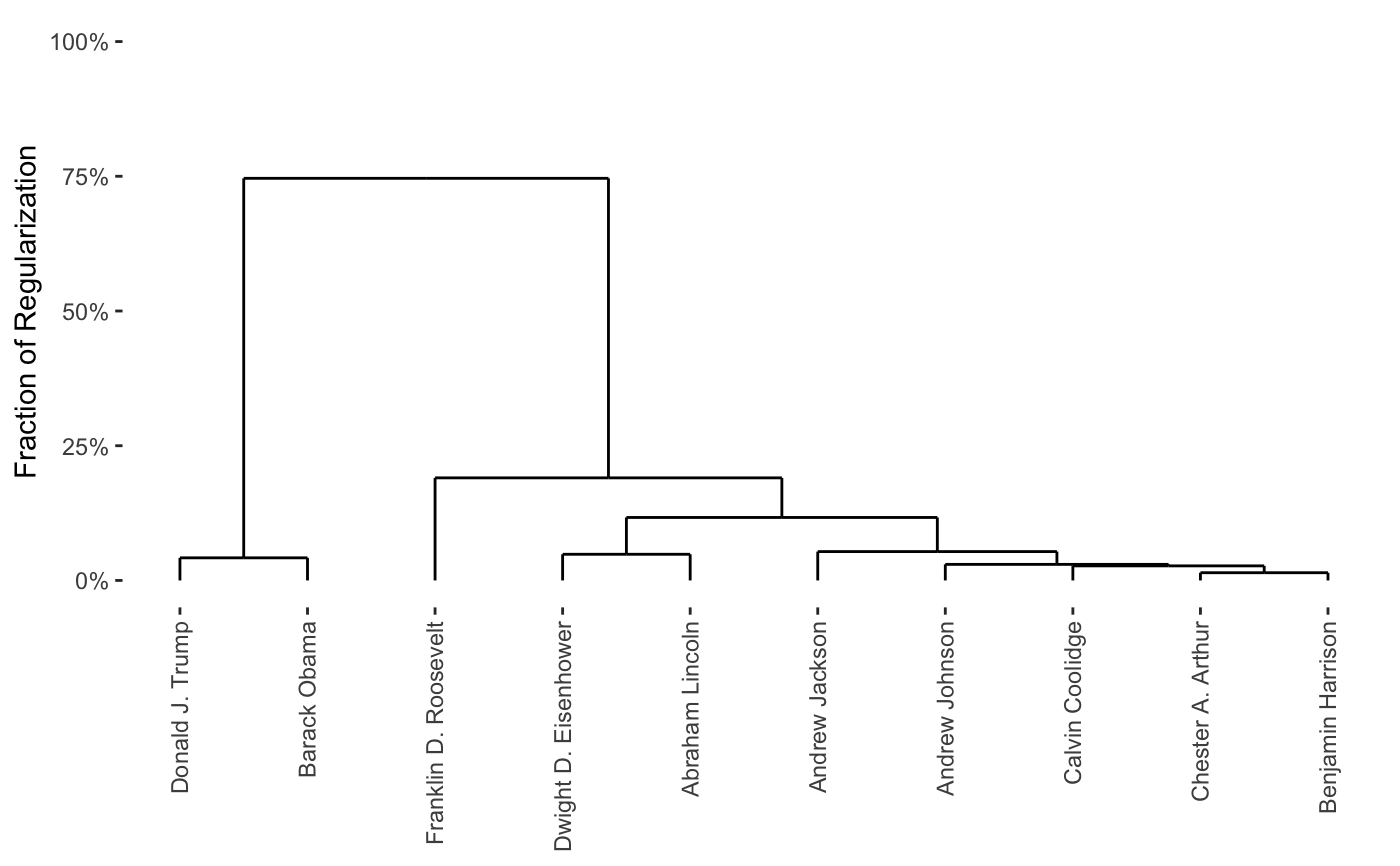
carp_fit <- CARP(presidential_speech[1:10,1:4])#>#>#>print(carp_fit)#> CARP Fit Summary #> ==================== #> #> Algorithm: CARP (t = 1.05) #> Fit Time: 0.008 secs #> Total Time: 1.025 secs #> #> Number of Observations: 10 #> Number of Variables: 4 #> #> Pre-processing options: #> - Columnwise centering: TRUE #> - Columnwise scaling: FALSE #> #> Weights: #> - Source: Radial Basis Function Kernel Weights #> - Distance Metric: Euclidean #> - Scale parameter (phi): 0.1 [Data-Driven] #> - Sparsified: 2 Nearest Neighbors [Data-Driven] #>plot(carp_fit)