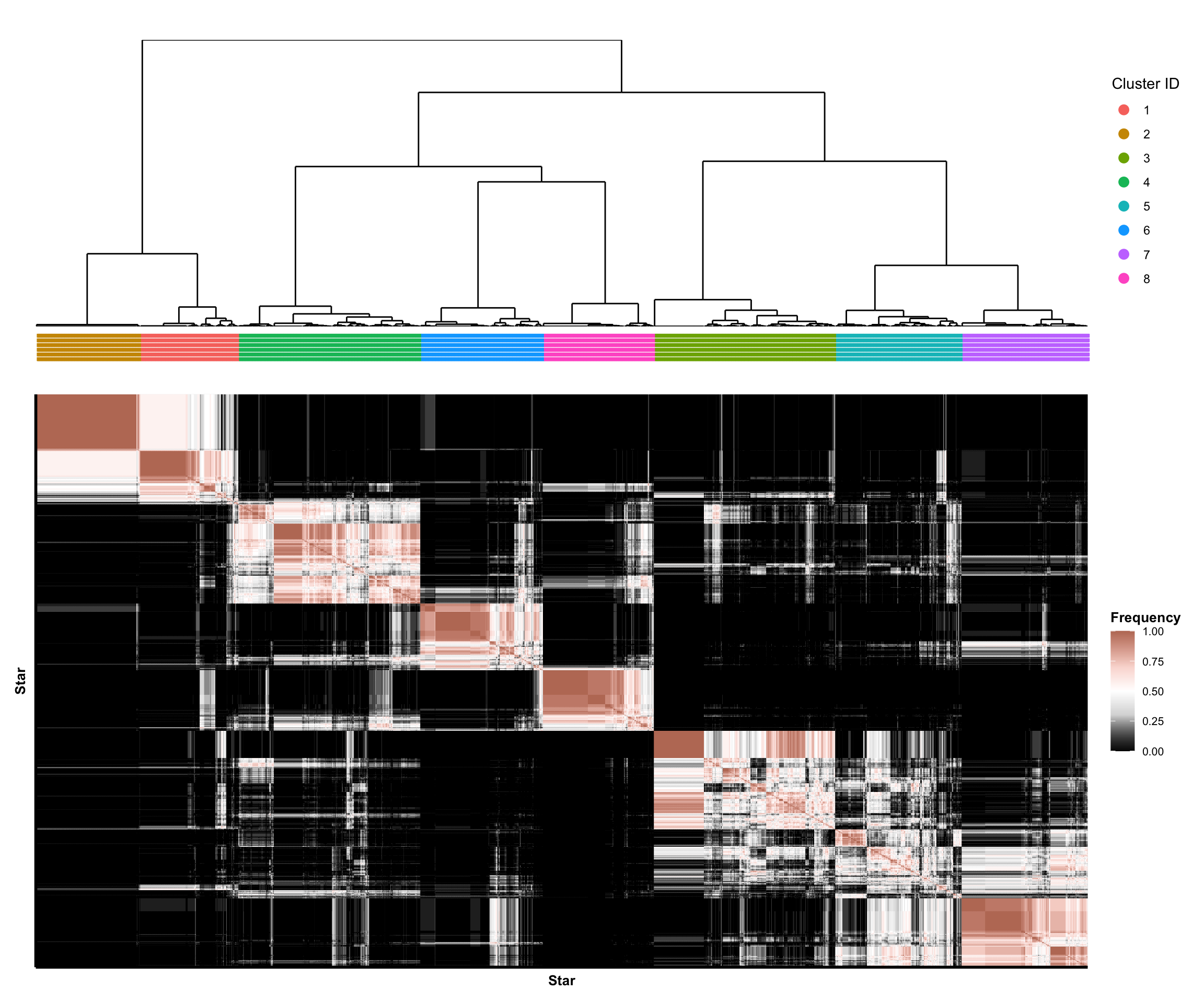
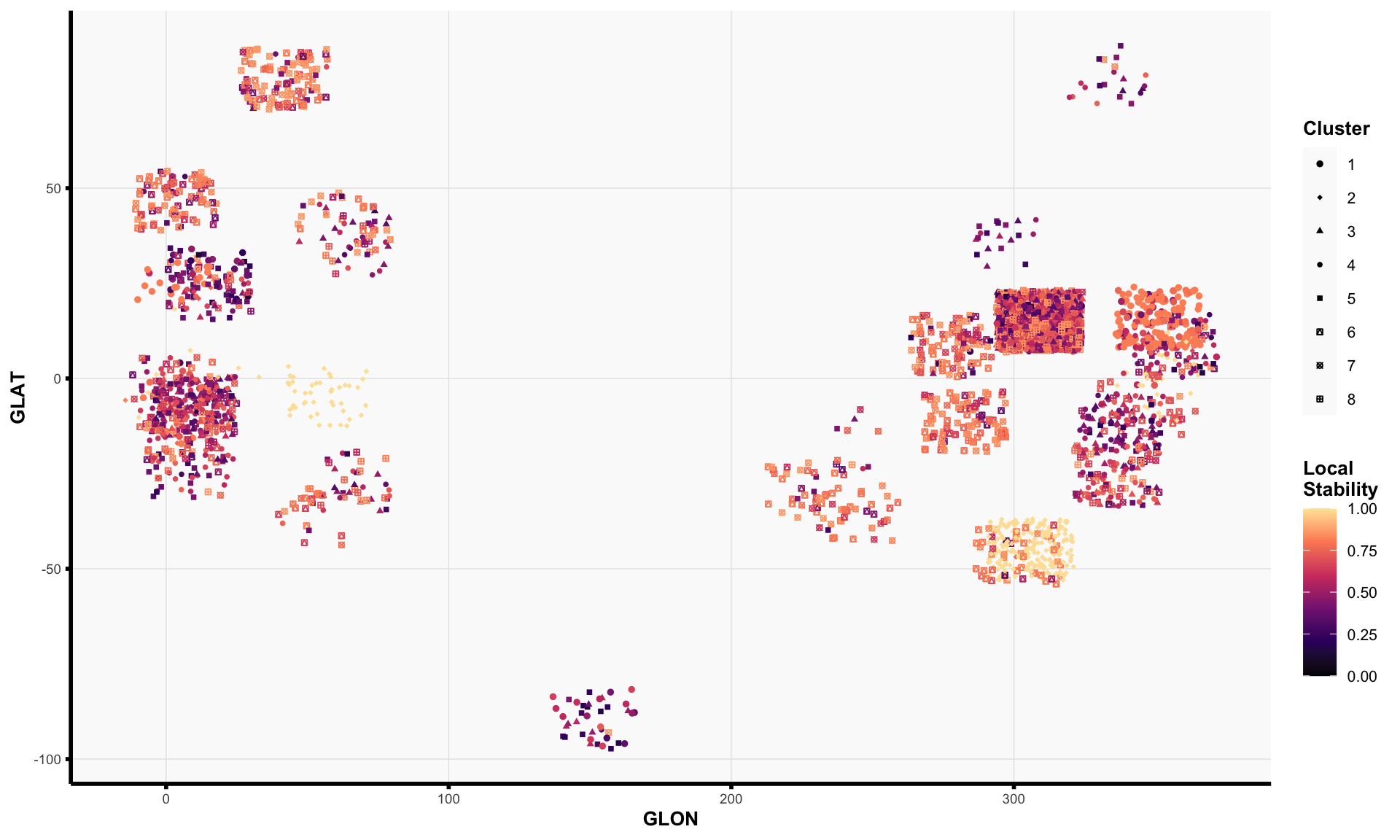
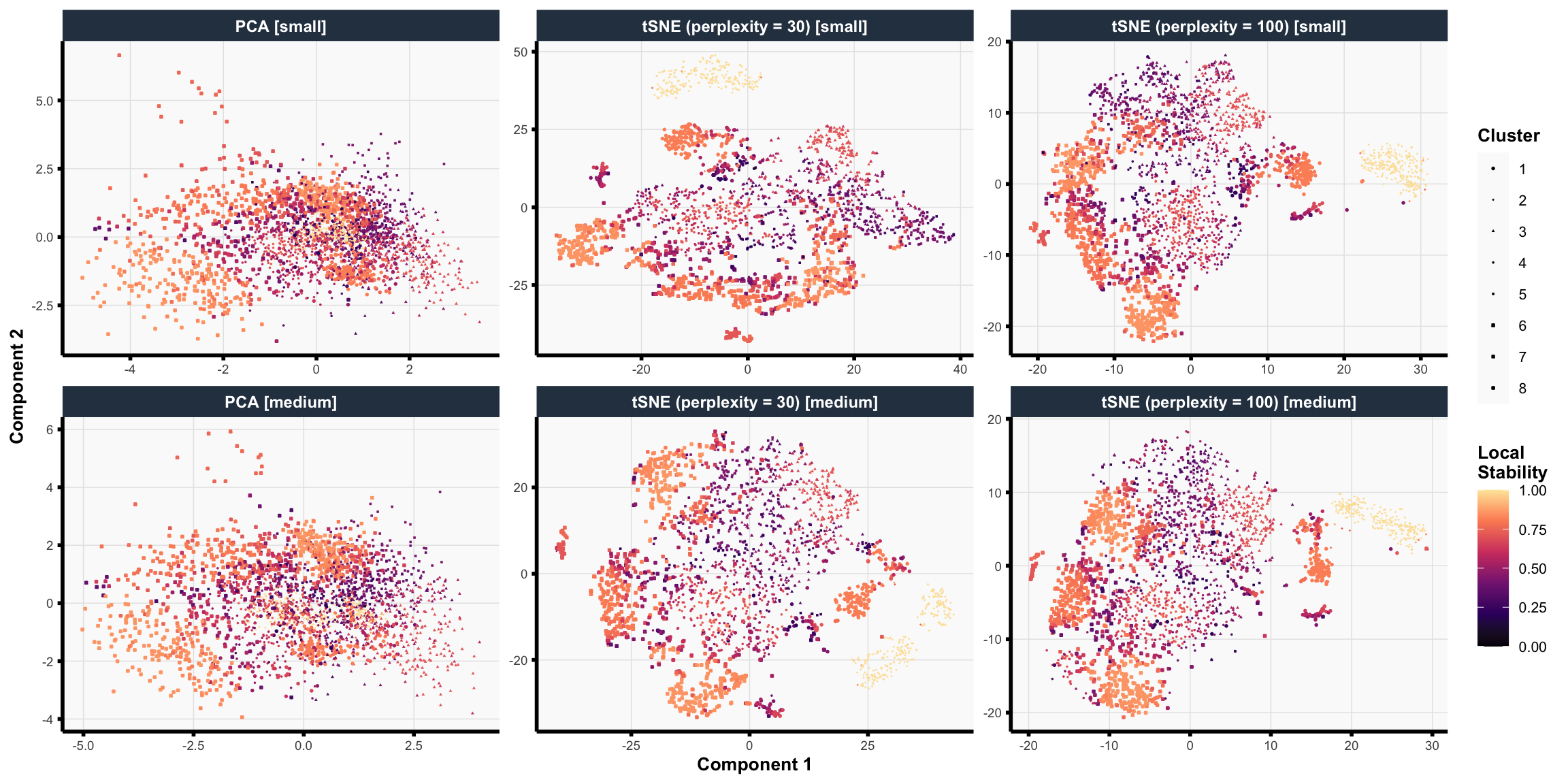
## this code chunk fits the final (tuned) clustering pipeline on the full data #### choose imputation methods #### <- list ("Mean-imputed" = rbind (data_mean_imputed$ train, data_mean_imputed$ test),"RF-imputed" = rbind (data_rf_imputed$ train, data_rf_imputed$ test)#### choose number of features #### <- list ("Small" = 7 ,"Medium" = 11 ,"Big" = 19 #### choose dimension reduction methods #### # raw data <- list ("Raw" = function (x) x)# pca <- list ("PCA" = purrr:: partial (fit_pca, ndim = 4 ))# tsne <- c (30 , 100 )<- purrr:: map (~ purrr:: partial (fit_tsne, dims = 2 , perplexity = .x)|> setNames (sprintf ("tSNE (perplexity = %d)" , tsne_perplexities))# putting it together <- c (#### choose clustering methods #### # kmeans <- list ("K-means" = purrr:: partial (fit_kmeans, ks = ks))# spectral clustering <- c (60 , 100 )<- purrr:: map (~ purrr:: partial (ks = ks,affinity = "nearest_neighbors" ,n_neighbors = .x|> setNames (sprintf ("Spectral (n_neighbors = %s)" , n_neighbors))# putting it together <- c (#### Fit Clustering Pipelines #### <- tidyr:: expand_grid (data = data_ls,feature_mode = feature_modes,dr_method = dr_fun_ls,clust_method = clust_fun_ls|> :: mutate (impute_mode_name = names (data),feature_mode_name = names (feature_mode),dr_method_name = names (dr_method),clust_method_name = names (clust_method),name = stringr:: str_glue ("{clust_method_name} [{impute_mode_name} + {feature_mode_name} + {dr_method_name}]" |> # remove some clustering pipelines to reduce computation burden :: filter (# remove all big feature set + dimension-reduction runs ! ((dr_method_name != "Raw" ) & (feature_mode_name == "Big" )),# restrict to tuned models == !! best_clust_method_name<- split (pipe_tib, seq_len (nrow (pipe_tib))) |> setNames (pipe_tib$ name)<- file.path (RESULTS_PATH, "clustering_fits_final.rds" )<- file.path ("consensus_clusters_final.rds" <- file.path ("consensus_neighborhood_matrices_final.rds" if (! file.exists (fit_results_fname) || ! file.exists (consensus_clusters_results_path) || ! file.exists (consensus_nbhd_results_path)) {library (future)plan (multisession, workers = NCORES)# fit clustering pipelines (if not already cached) <- furrr:: future_map (function (pipe_df) {<- create_preprocessing_pipeline (feature_mode = pipe_df$ feature_mode[[1 ]],preprocess_fun = pipe_df$ dr_method[[1 ]]<- pipe_df$ clust_method[[1 ]](data = pipe_df$ data[[1 ]], preprocess_fun = greturn (clust_out).options = furrr:: furrr_options (seed = TRUE , globals = list (ks = best_k,create_preprocessing_pipeline = create_preprocessing_pipeline,get_abundance_data = get_abundance_data,tsne_perplexities = tsne_perplexities,n_neighbors = n_neighbors,fit_kmeans = fit_kmeans,fit_spectral_clustering = fit_spectral_clustering# save fitted clustering pipelines saveRDS (clust_fit_ls, file = fit_results_fname)# estimate consensus clusters <- purrr:: map (clust_fit_ls, ~ .x$ cluster_ids) |> :: list_flatten (name_spec = "{inner}: {outer}" )<- get_consensus_neighborhood_matrix (clust_fit_ls)<- fit_consensus_clusters (nbhd_mat, k = best_k)saveRDS (consensus_out, file = consensus_clusters_results_path)saveRDS (nbhd_mat, file = consensus_nbhd_results_path)else {# read in results (if already cached) <- readRDS (fit_results_fname)<- readRDS (consensus_clusters_results_path)<- readRDS (consensus_nbhd_results_path)